Background :
TM-Aligner is multiple sequence alignment tool for Transmembrane proteins, TM-Aligner homepage provides an easy access point for performing sequence alignment.
TM-Aligner Homepage:
Figure 1. TM-Aligner Homepage.
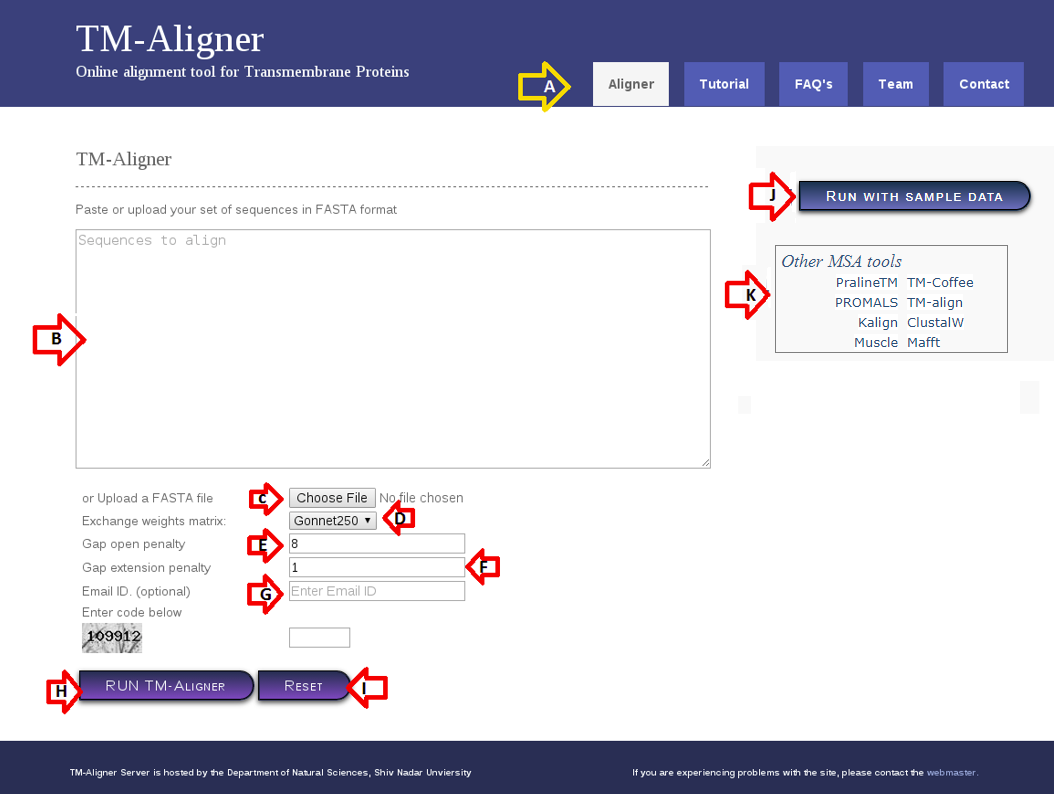
TM-Aligner homepage consists of several section each performs a specific set of function:
1). Common header (A) helps user to navigate through web server.
2). Input sequences are either pasted in the Text-area provided at (B), or can be uploaded using 'Choose file' option at (C). Remember input sequences should be in fasta format without special characters, numbers and non-amino acid characters. Input file/seq are checked for number of sequences (~5000) and size (~2MB) before submitting to server (for aligning more than 5000 sequences please contact webmaster). TM-Aligner accepts ".fa/.fasta/.txt/.afasta" file formats only.
3). TM-Aligner offers selection of the available Residue Exchange Matrices for scoring the alignments, including Dayhoff's PAM250 log odds matrix, BLOSUM62, PHAT (Default) a transmembrane-specific substitution matrix and GONNET250, user can select Exchange weights matrix using drop-down box provided in section (D).
4). User can also change Gap opening penalty (Default:8) and Gap Extension(Default:1) penalties from section (E) and (F).
5). User can specify email id at section (G) which will be used for forwarding result after job completion.
6). "RUN TM-Aligner" button is being provided at (H) for submitting sequences to server. User can clear form using "RESET" button at (I).
7). Section (J) contain button to run TM-Aligner with sample data with default parameters (Exchange matrix : PHAT, Gap Open penalty 8 & Gap Extension penalty 1). Section (K) contain link to sample data.
8). Links to other alignment tools are provided in section (L).
Get your alignment
Depending how complex is users input sequences the alignment can take sometime to get completed.You need to keep your browser open to wait for the result.
If you have entered your email address in your request you will be notified in your inbox when the alignment is complete.
When the alignment is complete it will displayed a page like the following.
The alignment result will be available on our server for seven days since your last access to it. Result link will be send to users account.
Figure 2. TM-Aligner Result-page
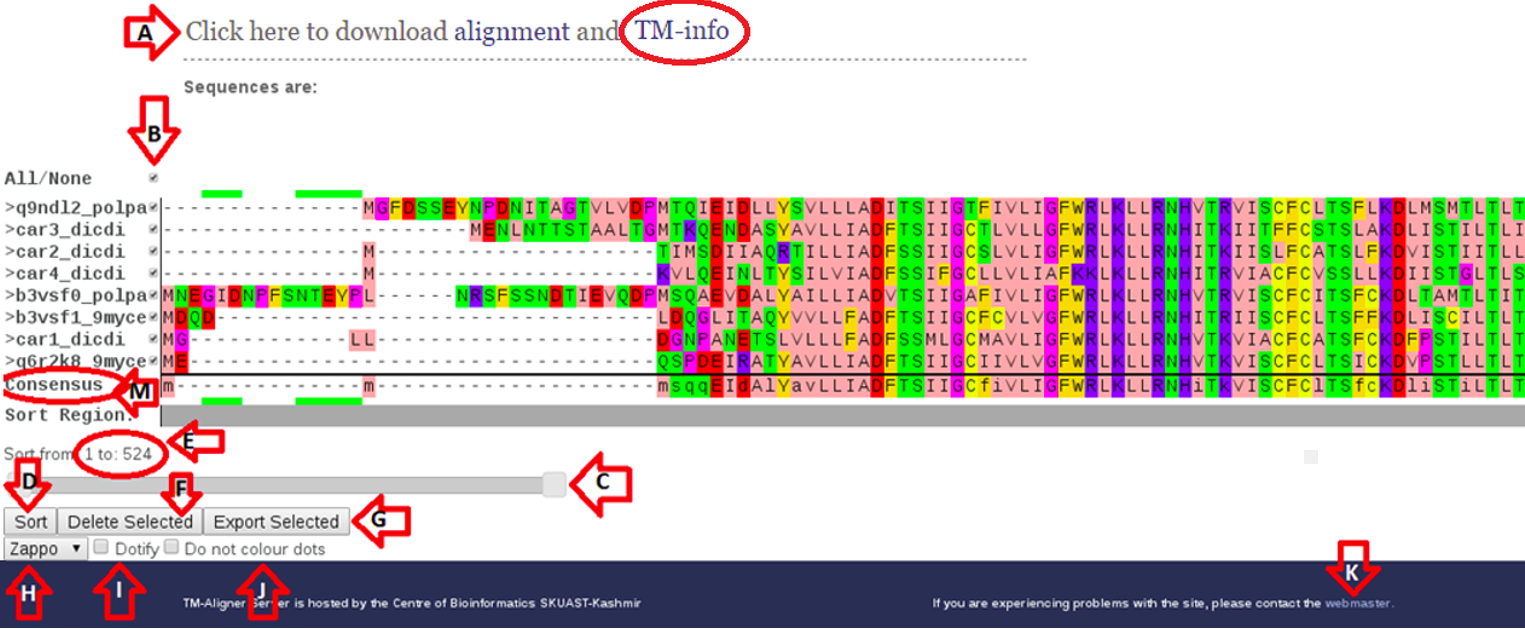
1).Link is provided at section (A) to download result and visualization/downloading transmembranes.
2). Check-boxes are provided at (B) to select/de-select sequences either to download, delete or export sequences to other server(s) using (F) and (G).
3). Sequence portion or complete sequence is selected using slider provided at (C) to sort sequences, residues selected for sorting are shown at (E) and sorting is being performed using (D).
4). Alignment visualization schema can be changed with the help of drop-down box available at section (H). For better visualization check-boxes at (I) and (J) are provided to replace repeated residues with dots and to change coloring schema.
5). Direct link to webmaster is given in section (K).
TM-info
Figure 3. TM-Info page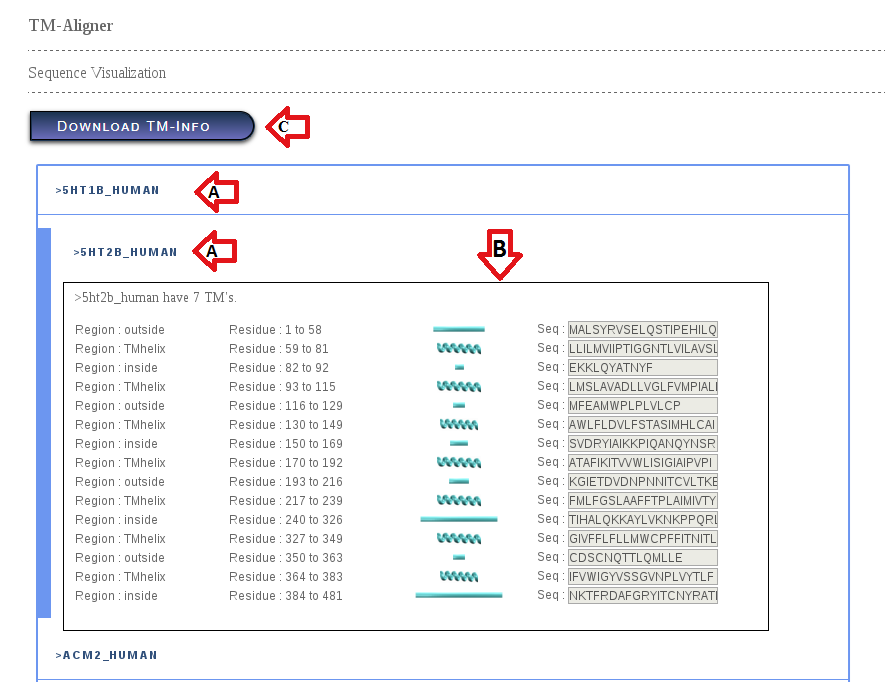
1). Input sequences goes at section (A).
2). Section (B) provides meta info about input seq(s) like location of TM region, TM seq etc
3). User can download TM - Info in MS - Excel from section (C).
Click here to download this tutorial in PDF format.